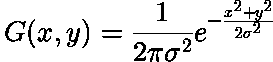Classical Computer Vision Basics¶
- Image representation
- Convolutional filters
- Thresholding
- Morphological operations
- erosion, dilation, etc.
- Instance segmentation
- connected components
- watershed
- Object analysis
What is Computer Vision?¶
- Computer Vision: "is an interdisciplinary scientific field that deals with how computers can gain high-level understanding from digital images or videos."
- Related field: signal processing: processing of (1d) digital signals
- many approaches can be extended to 2d (images) or higher d
- More general: also analysis of image series
- over time: videos
- over space: volumetric imaging (e.g. LIDAR, microscopy, medical imaging (CT-scans))
# Important python libraries:
import numpy as np # we always need numpy
import scipy.ndimage # image processing module in scipy
import matplotlib.pyplot as plt # for displaying images
import imageio # reading and writing images
import skimage # computer vision library
# import opencv <- also very popular, but harder to use
Image representation¶
- "Natural images": images of the natural world
- digital images are taken by digital camera (e.g. CCD)
- 2 image dimensions, 1 channel dimension (color space):
- H x W x C
- C = 3 (because of how we perceive color)
RGB representation¶
- Image has 3 color channels -> 3d "tensor"
- Dim 0, 1 = image dimensions, dim 2 = color channel
# look at an example rgb image
image = skimage.data.astronaut()
print(image.shape)
print(image.dtype, image.min(), image.max())
(512, 512, 3) uint8 0 255
fig, ax = plt.subplots(1, figsize=(8, 8))
ax.imshow(image)
ax.axis("off")
plt.show()
Other representations¶
RGB is not the only way to represent color space. Examples:
- CMYK: used for printing, because it's better applicable to ink mixing
- YUV: used for image compression
- HSV: Hue Saturation Value: used by artists, more natural to think about
# convert image to hsv and increase the saturation
image_hsv = skimage.color.rgb2hsv(image)
image_hsv[..., 1] = np.clip(image_hsv[..., 1] * 5, 0, 255)
more_saturation = skimage.color.hsv2rgb(image_hsv)
fig, ax = plt.subplots(1, 2, figsize=(8, 4))
ax[0].imshow(image)
ax[0].axis("off")
ax[1].imshow(more_saturation)
ax[1].axis("off")
plt.show()
Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
Image data formats & compression¶
- JPEG: lossy compression based on discrete fourier transform and discarding high frequency
- PNG: lossless compression using gzip
- TIFF: more complex format that supports multiple images and volumetric data
- MPEG: video format including lossy video compression
(also: vector graphics, e.g. svg)
Scientific images¶
- 3 channels (C=3) is specific to natural images (because of our perceptual color space)
- scientific images often just contain only single channel and are displayed as grayscale
- similar to old black & white fotos
- but can also contain more channels, e.g.
- multiple spectra in astronomy
- multiple fluorescent dies in microscopy
# load image of nuclei for normal and apoptotic cells
# (apoptose = programmed cell death)
url = "https://s3.embl.de/ilastik-downloads/2d_cells_apoptotic_1channel.png"
image = imageio.imread(url)
print(image.shape)
(1024, 1344)
fig, ax = plt.subplots(1, figsize=(12, 12))
ax.imshow(image, cmap="gray")
ax.axis("off")
plt.show()
Convolutional filters¶
- Kernel approximation of functions that are convolved with image.
- Can implement smoothing, edge detectors, texture detectors, etc.
Difference to CNNs:
- "classical" convolutional filters: fixed kernel, e.g. to smooth the image
- CNN: composition of many learned kernels
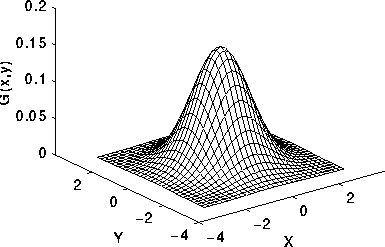
In practice: finite kernel approximation: (sigma=1)
# apply gaussian blur with 2 different sigma values
smoothed1 = skimage.filters.gaussian(image, 2)
smoothed2 = skimage.filters.gaussian(image, 8)
fig, ax = plt.subplots(1, 3, figsize=(16, 8))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[1].imshow(smoothed1, cmap="gray")
ax[1].axis("off")
ax[2].imshow(smoothed2, cmap="gray")
ax[2].axis("off")
plt.show()
Edge filter: Laplacian¶
Find edges in image using "Laplacian" filter (second spatial derivative):
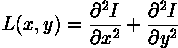
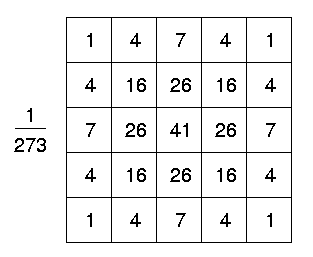
Kernel approximations:
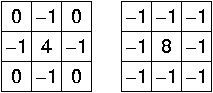
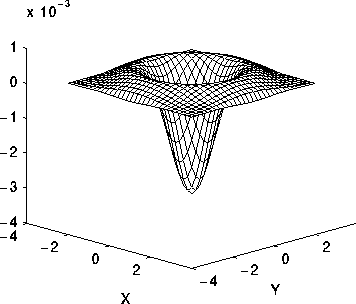
Problem: sensitive to noise => Apply gaussian smoothing first: Laplacian of Gaussian
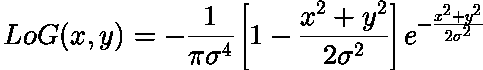
Profile of the Laplacian of Gaussian
# apply laplacian of gaussian filter to find edges
edges = skimage.filters.laplace(smoothed1)
fig, ax = plt.subplots(1, 2, figsize=(16, 8))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[1].imshow(edges, cmap="gray")
ax[1].axis("off")
plt.show()
More convolutional filters¶
- Difference of Gaussians: subtract two gaussians with different sigma -> edge filter
- Hessian Eigenvalues: eigenvalues of the matrix of sencond partial spatial derivatives -> texture filter
- Structure Tensor: measures gradient distribution -> texture filter
Thresholding¶
For analysis: segment image into different components (Semantic Segmentation)
simplest approach: foreground background segmentation through thresholding (Binary Segmentation)
Threshold can be derived from image intensity histogram

Advanced thresholding:
- Otsu's method to find threshold that separates foreground and background https://en.wikipedia.org/wiki/Otsu%27s_method
- Adaptive thresholding: find best threshold for local patch instead of one global threshold
# plot histogram of the image
fig, ax = plt.subplots(1, figsize=(8, 8))
histo = ax.hist(image.ravel())
plt.show()
# good threshold from histogram: 25-50
threshold = 50
# apply the threshold
thresholded = image > threshold
print("Values in thresholded image:", np.unique(thresholded))
fig, ax = plt.subplots(1, 2, figsize=(16, 8))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[1].imshow(thresholded)
ax[1].axis("off")
plt.show()
Values in thresholded image: [False True]
Morphological operations¶
Apply small structure element to image:
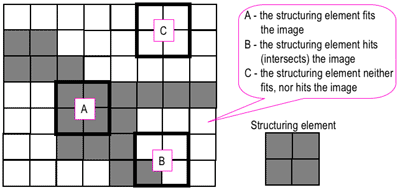
https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic4.htm
Erosion: Output is 1 if structure element fits the image -> shrinks the image
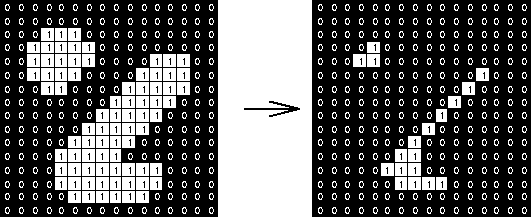
Dilation: Output is 1 if structure element hits the image grows the image
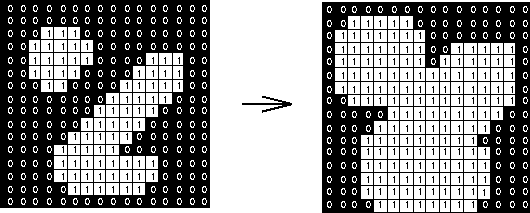
https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic4.htm
More complex morphological operations:
- opening: erosion followed by dilation: gets rid of small structures, large structures stay the same
- closing: dilation followed by erosion: fills holess
# apply erosion and dilation to our binary mask
eroded = scipy.ndimage.morphology.binary_erosion(thresholded, iterations=4)
dilated = scipy.ndimage.morphology.binary_dilation(thresholded, iterations=4)
fig, ax = plt.subplots(1, 3, figsize=(16, 16))
ax[0].imshow(thresholded)
ax[0].axis("off")
ax[1].imshow(eroded)
ax[1].axis("off")
ax[2].imshow(dilated)
ax[2].axis("off")
plt.show()
Instance Segmentation¶
Label each object in the image (and its pixels) with a unique id
- semantic segmentation:
- number of label values is known a priori (number of classes is known)
- instance segmentation:
- number of label values is not known a priori (number of objects is unknown)
Connected components¶
Label pixels that are connected in the image as one component Depends on the definition of neighborhood (i.e. connected or not)
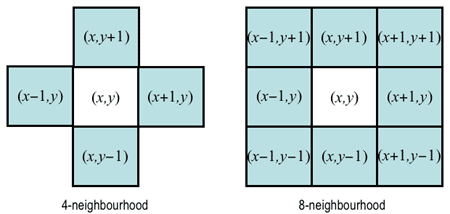
https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic3.htm#connect
Connected component labeling:
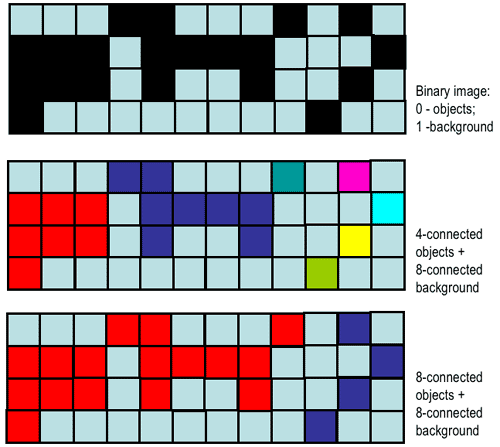
https://www.cs.auckland.ac.nz/courses/compsci773s1c/lectures/ImageProcessing-html/topic3.htm#connect
Connected Components Algorithm:
- uses Union Find Datastructure
- 2 Passes:
- first pass: find which pixels are connected
- second pass: assign to each pixel the label value of its component
# apply connected components to the foreground/background segmentation
components = skimage.measure.label(thresholded)
print("Number of components:", len(np.unique(components)))
Number of components: 227
# display the connected components using random colors (code not shown)
fig, ax = plt.subplots(1, 2, figsize=(16, 16))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[1].imshow(components, cmap=random_colors, interpolation="nearest")
ax[1].axis("off")
plt.show()
Watershed¶
Interpret image as height map; "flood" with water, watersheds correspond to object boundaries

Different perspective: Seeded watershed: grow regions starting from seeds:
Finding seeds:
- use local minima of height map
- or: use external markers
# find seeds, height map and mask for the watershed
# seeds: components of strong eroded threshold image
seeds = skimage.measure.label(
scipy.ndimage.binary_erosion(thresholded, iterations=8)
)
# height map: the edge filter
hmap = edges
# mask: the thresholded image
mask = thresholded
nuclei = skimage.segmentation.watershed(hmap, markers=seeds, mask=mask)
n_nuclei = len(np.unique(nuclei)) - 1
print("Number of segmented nuclei:", n_nuclei)
Number of segmented nuclei: 95
fig, ax = plt.subplots(1, 2, figsize=(16, 16))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[1].imshow(nuclei, cmap=random_colors, interpolation="nearest")
ax[1].axis("off")
plt.show()
Object analysis¶
Instance segmentation often is the basis for further object based analysis, e.g.
- count objects
- analyze spatial distribution
- compare morphology of the objects
- ...
Here: distinguish nuclei into normal and apoptotic based on mean object intensity
# compute the mean intensities per nucleus
nucleus_properties = skimage.measure.regionprops(nuclei, image)
mean_intensities = np.array([prop.mean_intensity for prop in nucleus_properties])
# look at the intensity histogram
h = plt.hist(mean_intensities)
apoptotic_threshold = 80
apoptotic_nuclei = np.where(mean_intensities >= 80)[0] + 1
normal_nuclei = np.where(mean_intensities < 80)[0] + 1
print("Nr normal nuclei:", len(normal_nuclei))
print("Nr apo nuclei:", len(apoptotic_nuclei))
Nr normal nuclei: 44 Nr apo nuclei: 51
# display the classified cells as image
cmap = colors.ListedColormap(["white", "blue", "yellow"])
fix, ax = plt.subplots(1, 2, figsize=(20, 10))
ax[0].imshow(image, cmap="gray")
ax[0].axis("off")
ax[0].set_title("Image")
ax[1].imshow(classified_nuclei, cmap=cmap, interpolation="nearest")
ax[1].set_title("Classified cells")
ax[1].axis("off")
(-0.5, 1343.5, 1023.5, -0.5)
# we arrived back at a semantic segmnentation!